Tularemia is a rare but often lethal disease. It is caused by one of the most aggressive pathogens on earth, the bacterium Francisella tularensis. The microbe, transported by a variety of animals and insects, is able to enter and attack the body through a range of pathways, resulting in different constellations of symptoms and degrees of severity.
Tularemia remains poorly understood and no safe and effective vaccine exists for the disease. The extreme lethality of F. tularensis and its potential to be aerosolized have also made it a bioweapon candidate, increasing the urgency of understanding the disease and developing effective treatments.
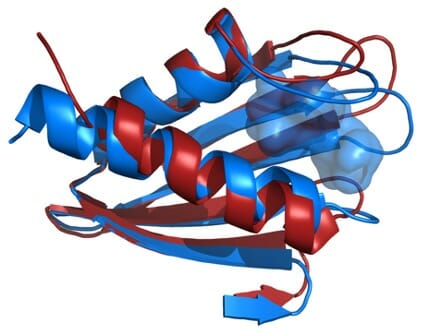
In a new study, researchers at the Biodesign Center for Applied Structural Discovery and the School of Molecular Sciences at ASU examine a key membrane protein responsible for the bacterium’s prodigious ability to infect the body and cause illness. This virulence factor, known as Flpp3, is examined in unprecedented detail with the aid of an X-ray free electron laser or XFEL, a massive and powerful X-ray accelerator located at the SLAC National Accelerator Laboratory, Stanford.
XFEL technology uses brilliant and extremely short flashes of X-rays to probe crystalized samples of Flpp3, revealing the protein’s detailed structure as never before. By comparing the structural information gathered by XFEL experiments with previous structural analysis using NMR, researchers have developed a more complete model of Flpp3’s elaborate form. (Previous studies have shown that when a gene coding for Flpp3 is disabled, the effects of the F. tularensis pathogen are significantly diminished.)
Scientists hope to eventually use this information to develop targeted drugs capable of disabling the protein’s virulence properties and protecting against tularemia. The advances in understanding could also help scientists develop an effective vaccine against the illness in the future. Currently, only live attenuated vaccines exist for tularemia, and risks of infection and insufficient immunity associated with this approach have precluded use of such vaccines in the US.

Biodesign researcher Dr. James Zook together with Professors Petra Fromme and Abhishek Singharoy at the Biodesign Center for Applied Structural Discovery led the new study. He is joined by international colleagues, including researchers from DESY, SLAC, AstraZeneca, the European Molecular Biology Laboratory in Grenoble, France, among others.
Petra Fromme, director of the Center for Applied Structural Discovery explains the Importance of the results: “This study combines, for the first time, state of the art XFEL techniques with NMR and molecular modelling to unravel the large conformational space of Flpp3. The study unravels different conformations of this important protein for the virulence of the bacterium in the XFEL and NMR structure thereby showing the highly dynamic nature of Flpp3. This study is so exciting as it shows that very different conformations coexist and are converted into each other under physiological conditions.”
Visualizing virulence
Using detailed structural data from NMR and new XFEL analysis of the tularemia virulence factor, the researchers identified a potential inhibitor of Flpp3. This information was obtained from available virtual libraries containing structures of drug fragments. Next, a physics-based modeling method, known as molecular dynamics (MD), provided detailed information on the fluctuations and conformational changes of atoms and molecules in the virulence-linked protein, helping researchers get a more precise read on Flpp3’s structure and behavior.
“This work provides several atomic-resolution structures of an important virulence factor from the bacterium that causes tularemia,” according to Biodesign researcher Dr.Debra Hansen, a co-author of the new study. The protein configurations identified will help researchers pursue structure-based design of drugs that could be effective against the elusive disease, through the targeting and inhibition of Flpp3.

As co-author and Biodesign researcher Dr. Abhishek Singharoy explains, the study is noteworthy for being among the first investigations of protein conformational flexibility discovered with serial femtosecond X-ray crystallography and NMR and confirmed using molecular dynamics (MD) simulations.
The group’s findings appear in the current issue of the Cell Press journal Structure.
Persistent threat
Tularemia, also known as rabbit fever, is a rare infectious disease, typically attacking the eyes, skin, lymph nodes and lungs following infection by the bacterium F. tularensis. The disease is endemic in North America as well as in many parts of Europe and Asia, though cases of tularemia are uncommon and full-blown outbreaks tend to be restricted to regions with poor sanitation and inaccessibility to modern healthcare.
Tularemia primarily affects mammals, particularly rodents, rabbits and hares, although it sometimes also infects birds, sheep, and domestic animals, including dogs, cats and hamsters.
The disease can be spread to humans through insect bites and direct exposure to an infected animal. The disease is extremely contagious. Just 10 bacterial cells can be fatal and a single bacterium may be sufficient to cause infection. The organism can live for weeks in soil, water and dead animals.
Tularemia can be effectively treated if detected early, though the treatment regime can be lengthy and complex. Most infected with F. tularensis display symptoms within three to five days, though it can take as long as two weeks.
The disease exists in a variety of forms with differing symptoms, depending on how and where the bacteria enter the body. These include ulceroglandular tularemia, the most common form, which produces ulcers of the skin at the site of infection, swollen and painful lymph glands, fever, chills, headache and exhaustion.
Other forms include glandular, oculoglandular, oropharyngeal, pneumonic, and typhoidal tularemia. If left untreated, a variety of severe complications from the disease may ensue, including meningitis, inflammation of the lungs, irritation around the heart and infection of bone.
Crystal method
In the current study, a technique known as serial femtosecond X-ray crystallography is used to probe the structure of the Flpp3 protein. Here, brief and brilliant X-ray bursts, roughly a billion times brighter than conventional X-rays, strike a jet of crystals “flying” through the X-ray beam. The intense X-ray blast destroys the crystals but before doing so, creates a diffraction pattern on a screen. The X-ray pulses are ultrashort, lasting just 40 femtoseconds, that they outrun X-ray damage, allowing for data to be collected at room temperature under near physiological conditions. (1 fs = 10-15 seconds or one quadrillionth of a second.)
Assembling many of these X-ray shots with the aid of computers permits the assembly of a detailed, 3D structure of the protein under study. This so-called diffraction before destruction method was first pioneered by Henry Chapman at the Deutsches Elektronen-Synchrotron (DESY) with the team at ASU under the lead of John Spence and Petra Fromme and their collaborators.
The researchers combined the new XFEL structural data with their previous NMR studies of Flpp3, observing two distinct states of the protein. The MD simulations revealed an internal cavity structure that is transient, suggesting that Flpp3 undergoes a subtle conformational change.
The approach opens the door to targeted drug development aimed at reducing the lethality of tularemia and displays the power of combined technologies for unlocking the details of protein structure and dynamics.
In addition to their positions at the Biodesign institute, Petra Fromme, Debra Hansen and Abhishek Singharoy are researchers at ASU’s School of Molecular Sciences.
This research used resources of the Oak Ridge Leadership Computing Facility, which is supported by the Office of Science, Department of Energy (DE-AC05-00OR22725).Use of LCLS, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. Parts of the sample injector used at LCLS for this research were funded by the NIH, P41GM103393, formerly P41RR001209. This work was supported by the NIH Common Fund Grant R01 GM095583 (to P.F.) and the Center for Membrane Proteins in infectious diseases, MPID supported by the PSI:Biology Structural Biology initiative of the NIH U54 GM094599. We also acknowledge funding from the Science and Technology Center (STC) “BioXFEL” through award STC-1231306. The work was also supported by funds from the Biodesign Center for Applied Structural Discovery at Arizona State University.